
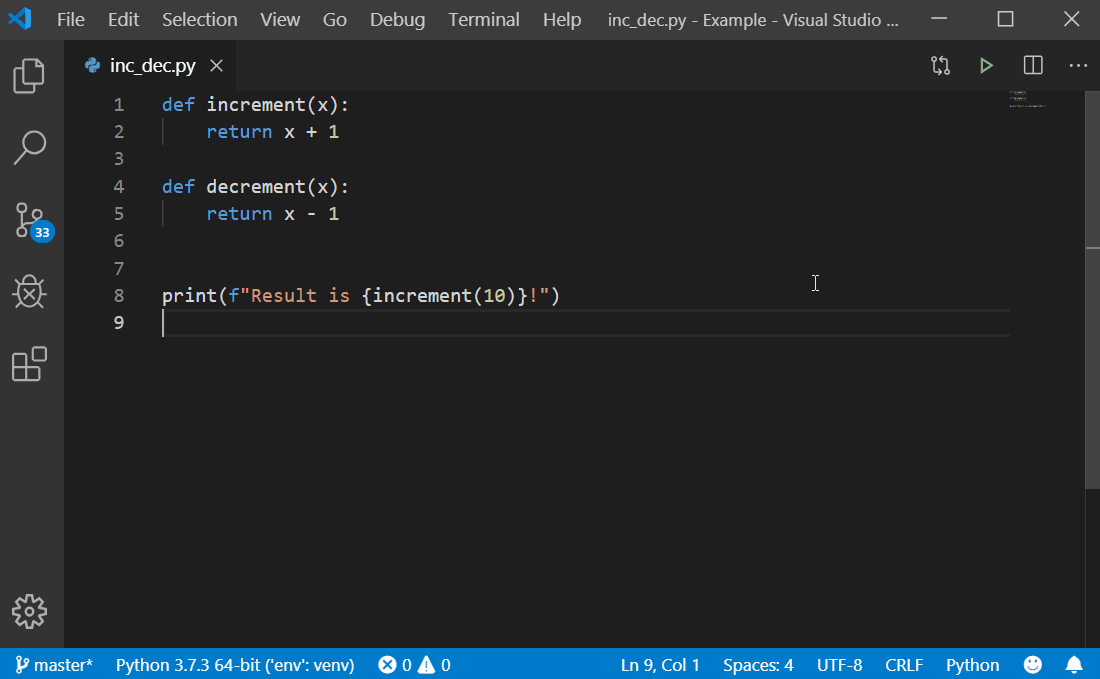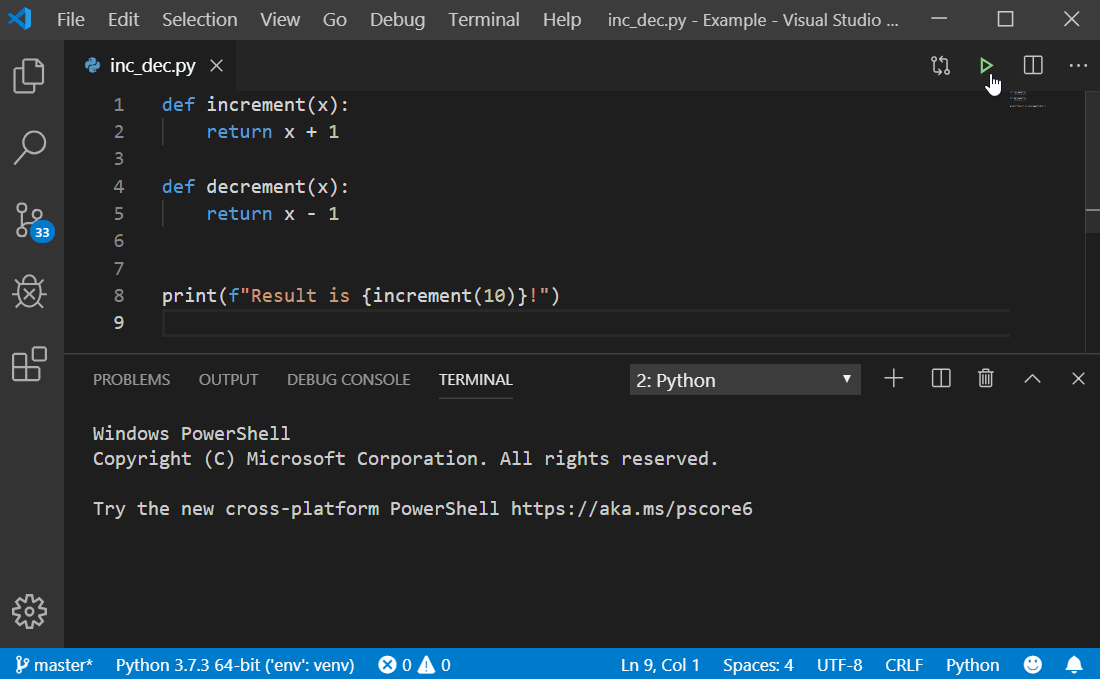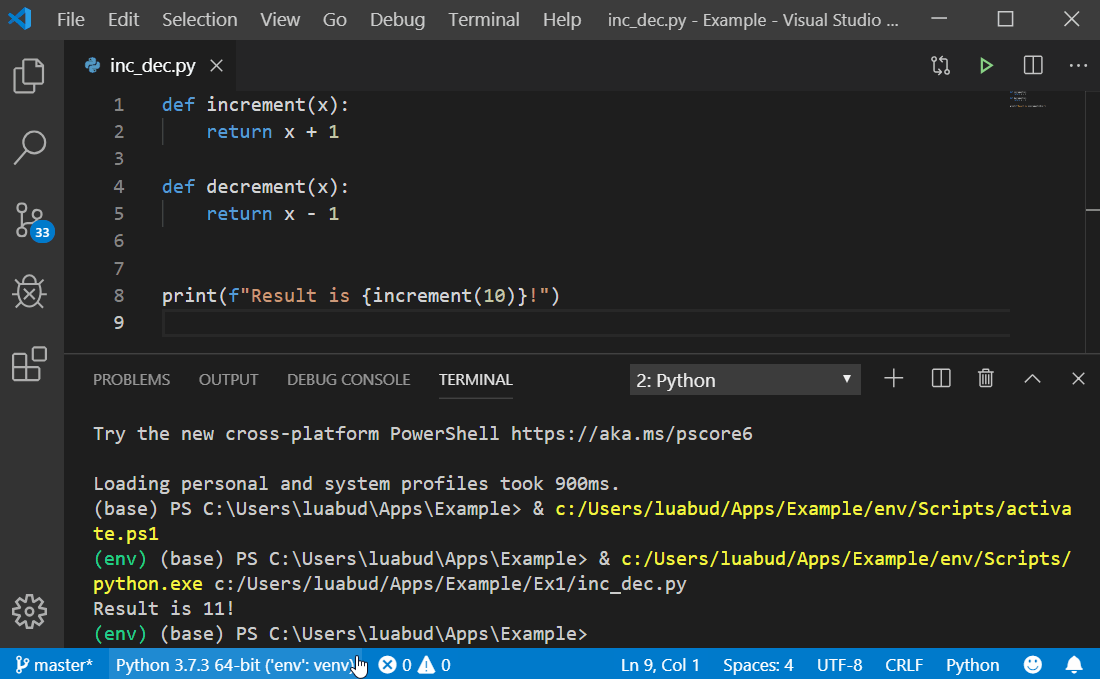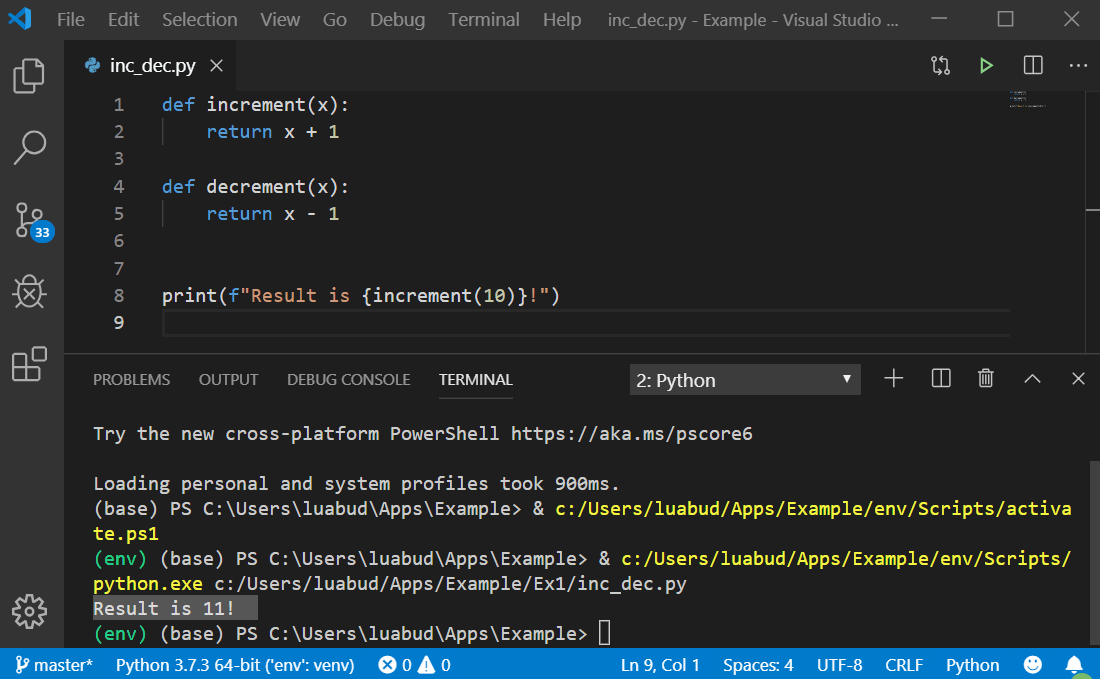
#Run a python script from terminal install
The recommended approach is to (1) keep a single JupyterLab installation in the base conda environment and (2) install a Python kernel separately in the conda environment of the desired project i.e. /rebates/2ftutorial2frunning-a-python-script&. While we could also install JupyterLab in the current ipp_analysis conda environment, this approach can become inconvenient as we install more conda environments in the future. We’ve already installed JupyterLab in the base conda environment. This will execute a script and put all its global variables in the interpreters global scope (the normal behavior in most scripting environments). EDIT: Im trying to use variables and settings from that file, not to invoke a separate process. Next, we’ll explore how JupyterLab allows us to execute Python code via a user-friendly web-based interface. Im trying to execute a file with Python commands from within the interpreter. To run Python from a terminal on Windows: Click the search icon (magnifying glass) in the bottom-left corner of the screen. So far we’ve seen how Python code can be executed via the command-line terminal interface. P.S find a screenshot of my terminal window. Im running spark in windows 64bit architecture system with JDK 1.8 version.
 The command is, spark-submit -master
The command is, spark-submit -master To know more about IPython, you may visit their official website. Spark environment provides a command to execute the application file, be it in Scala or Java(need a Jar format), Python and R programming file. In addition, IPython extends the basic Python Shell with a rich set of features and makes it more powerful. IPython can execute anything that can be executed in the basic Python Shell.


 0 kommentar(er)
0 kommentar(er)
